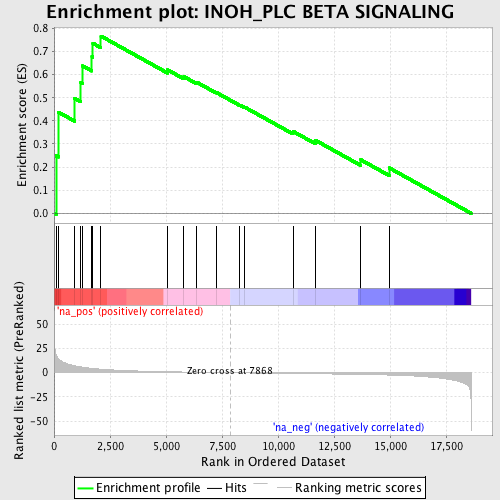
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | INOH_PLC BETA SIGNALING |
| Enrichment Score (ES) | 0.7667094 |
| Normalized Enrichment Score (NES) | 1.8685412 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.03957353 |
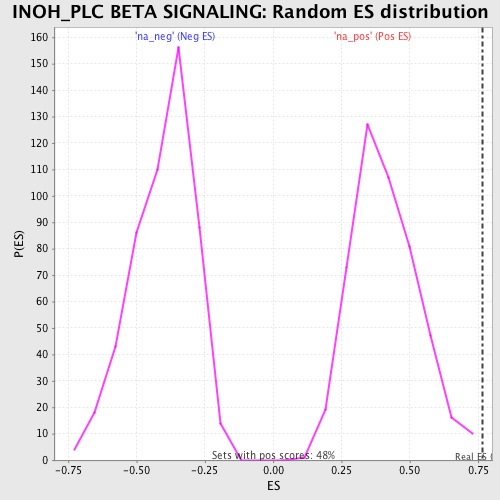
| FWER p-Value | 0.272 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PRKCH | 92 | 18.399 | 0.2491 | Yes | ||
| 2 | PRKCQ | 196 | 14.046 | 0.4376 | Yes | ||
| 3 | GNAQ | 918 | 7.135 | 0.4973 | Yes | ||
| 4 | ITPR1 | 1193 | 5.947 | 0.5647 | Yes | ||
| 5 | PRKCD | 1279 | 5.615 | 0.6377 | Yes | ||
| 6 | MAPK3 | 1689 | 4.468 | 0.6774 | Yes | ||
| 7 | PLCB3 | 1732 | 4.377 | 0.7356 | Yes | ||
| 8 | ITPR3 | 2092 | 3.648 | 0.7667 | Yes | ||
| 9 | MAP2K1 | 5069 | 1.006 | 0.6206 | No | ||
| 10 | RAF1 | 5794 | 0.702 | 0.5914 | No | ||
| 11 | ARAF | 6377 | 0.479 | 0.5667 | No | ||
| 12 | PLCG1 | 7237 | 0.186 | 0.5231 | No | ||
| 13 | PRKCI | 8291 | -0.129 | 0.4682 | No | ||
| 14 | PRKCA | 8501 | -0.195 | 0.4597 | No | ||
| 15 | PRKCZ | 10666 | -0.815 | 0.3546 | No | ||
| 16 | PRKCE | 11664 | -1.095 | 0.3161 | No | ||
| 17 | PLCB1 | 13693 | -1.764 | 0.2314 | No | ||
| 18 | MAP2K2 | 14962 | -2.404 | 0.1965 | No |